Appendix C — Chapter 4 supplemental material
You are reading the work-in-progress of this thesis.
This chapter should be readable but is currently undergoing final polishing.
C.1 Data wrangling
The data wrangling processes were performed using the targets R package. The full pipeline can be seen in the _targets.R file at the root of the research compendium.
C.2 Distribution of main variables
source(here::here("R/test_normality.R"))
source(here::here("R/utils.R"))
col <- "age"
stats <- data |>
magrittr::extract2(col) |>
test_normality(
name = col,
threshold = hms::parse_hms("12:00:00"),
remove_outliers = FALSE,
iqr_mult = 1.5,
log_transform = FALSE,
density_line = TRUE,
text_size = env_vars$base_size,
print = TRUE
) |>
rutils:::shush()
#> # A tibble: 14 × 2
#> name value
#> <chr> <chr>
#> 1 n 79198
#> 2 n_rm_na 79198
#> 3 n_na 0
#> 4 mean 31.9838074965417
#> 5 var 85.2414919292643
#> 6 sd 9.23263190695179
#> # ℹ 8 more rows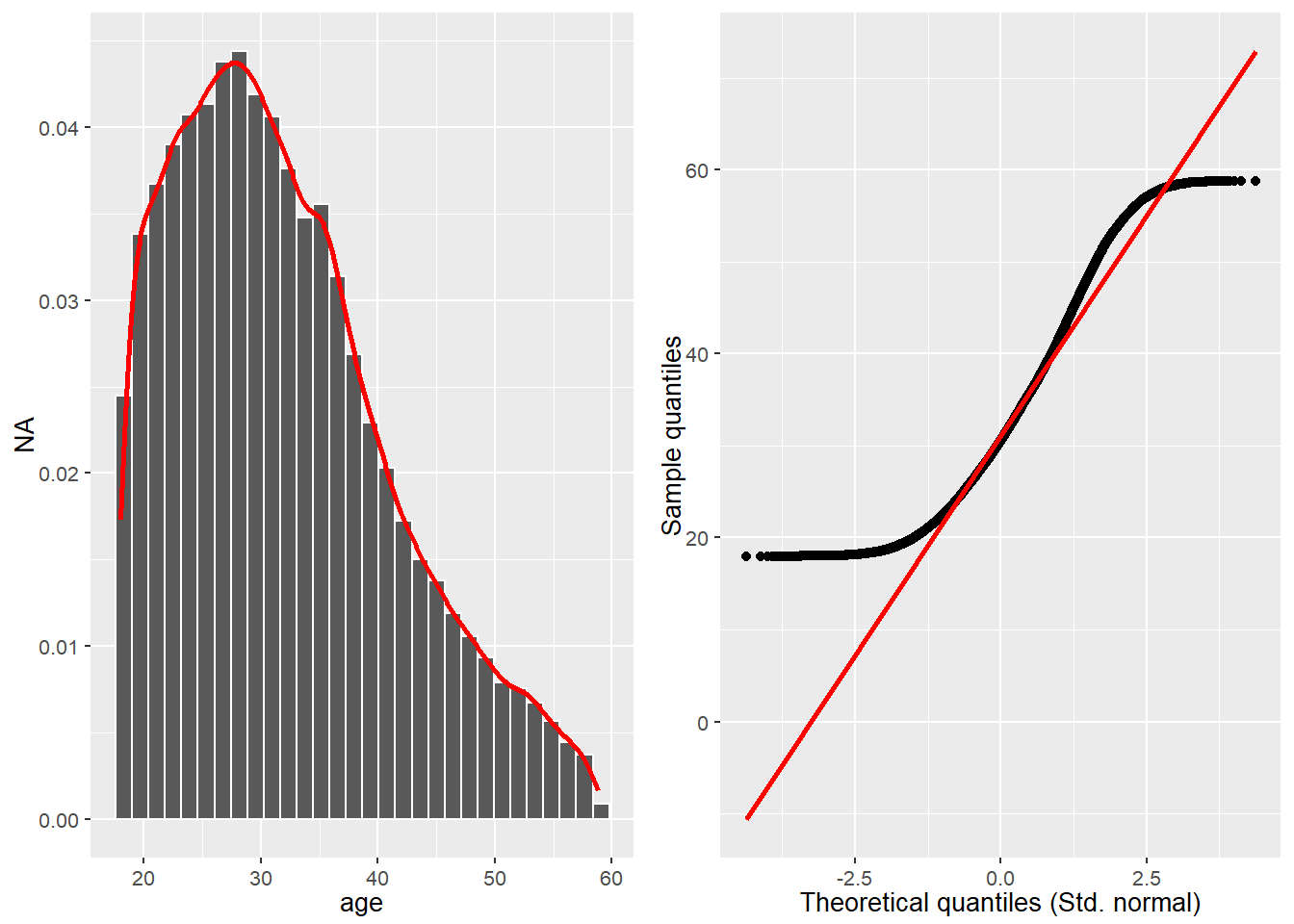
Source: Created by the author.
stats$stats |> list_as_tibble()| name | value |
|---|---|
| n | 79198 |
| n_rm_na | 79198 |
| n_na | 0 |
| mean | 31.9838074965417 |
| var | 85.2414919292643 |
| sd | 9.23263190695179 |
| min | 18 |
| q_1 | 24.7222222222222 |
| median | 30.5388888888889 |
| q_3 | 37.61875 |
| max | 58.7861111111111 |
| iqr | 12.8965277777778 |
| skewness | 0.665751526654394 |
| kurtosis | 2.82381488030798 |
Source: Created by the author.
C.3 Geographic distribution
source(here::here("R/plot_brazil_uf_map.R"))
rutils:::assert_internet()
brazil_uf_map <-
data |>
plot_brazil_uf_map(option = "viridis", text_size = env_vars$base_size)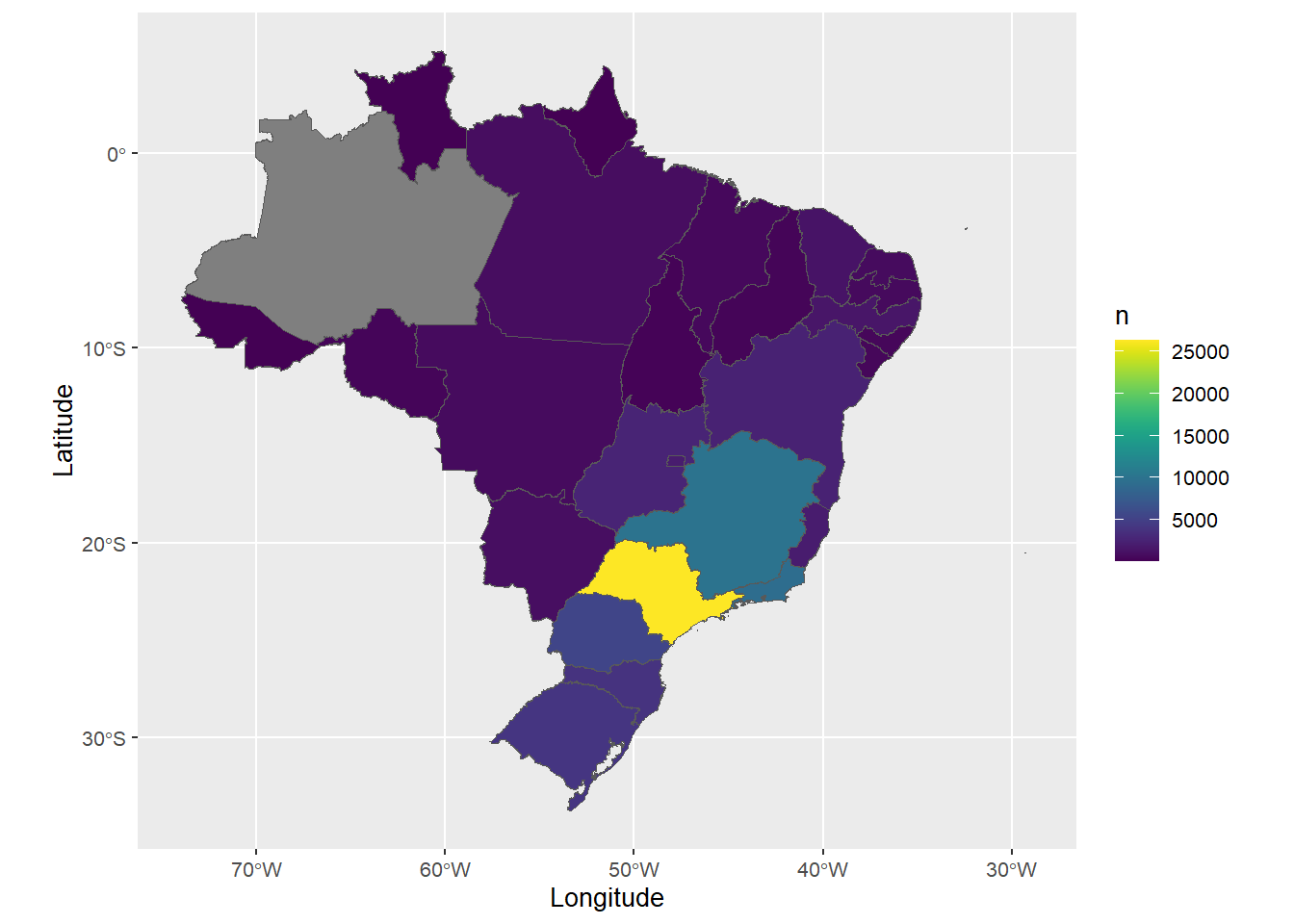
Source: Created by the author.
C.4 Age pyramid
source(here::here("R/plot_age_pyramid.R"))
age_pyramid <-
data |>
plot_age_pyramid(
interval = 10,
na_rm = TRUE,
text_size = env_vars$base_size
)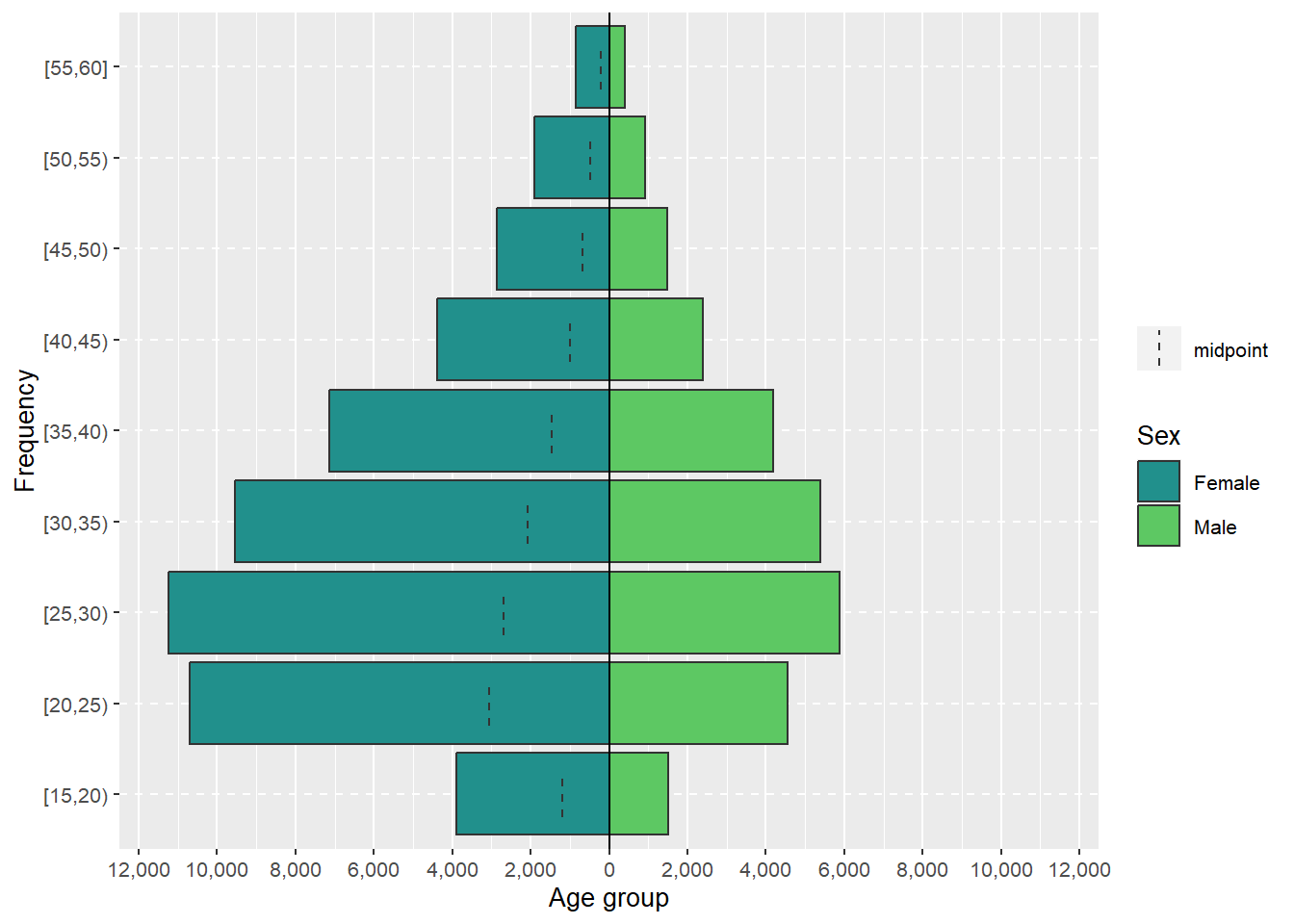
Source: Created by the author.
C.5 Correlation matrix
source(here::here("R/plot_ggcorrplot.R"))
cols <- c("sex", "age", "latitude", "longitude", "msf_sc", "sjl", "le_week")
ggcorrplot <-
data |>
plot_ggcorrplot(
cols = cols,
na_rm = TRUE,
text_size = env_vars$base_size,
hc_order = TRUE
)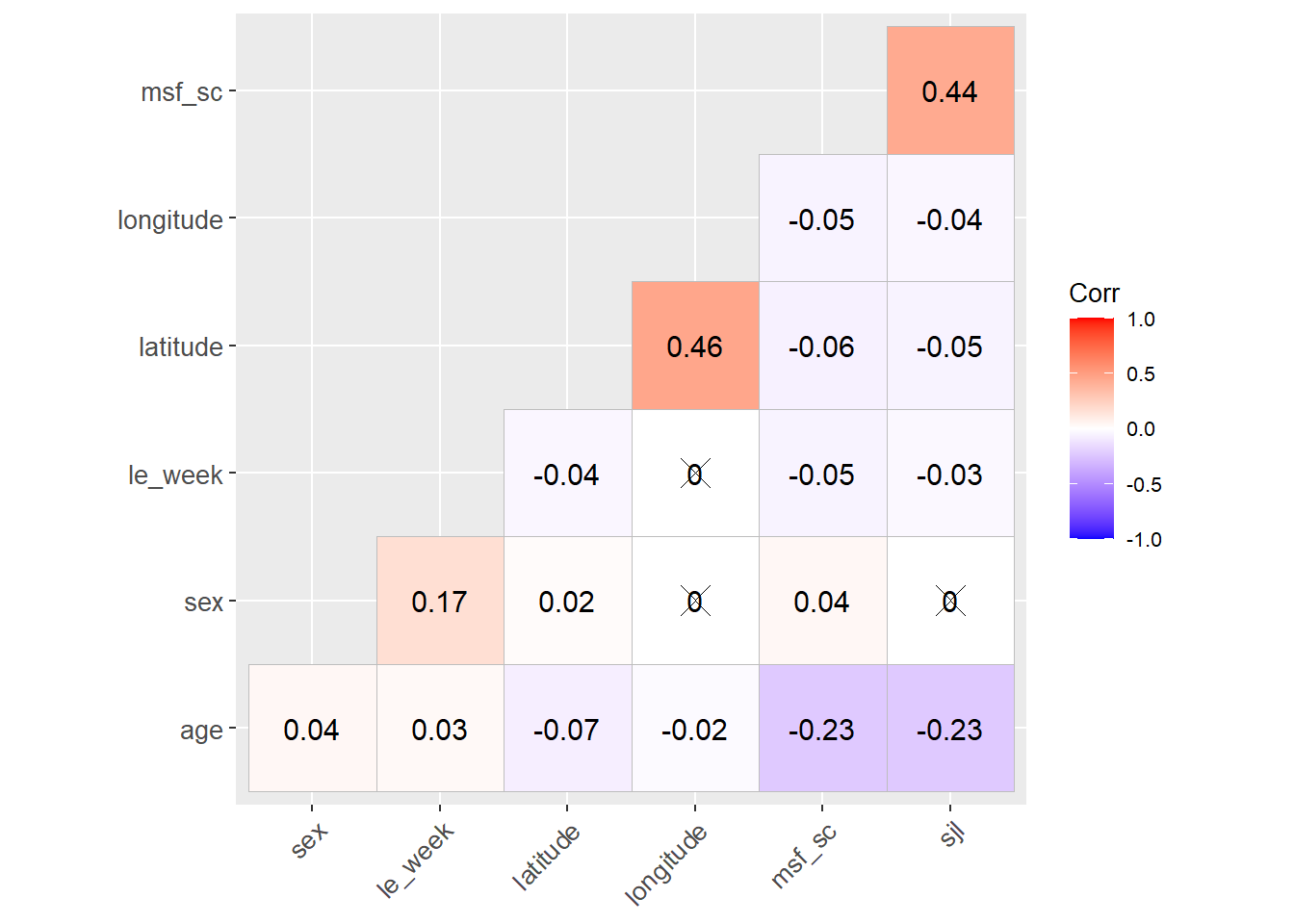
Source: Created by the author.
C.6 Age and sex series
C.6.1 Age/sex versus chronotype
source(here::here("R/plot_age_series.R"))
col <- "msf_sc"
y_lab <- latex2exp::TeX("Local time ($MSF_{sc}$)")
data |>
dplyr::filter(age <= 50) |>
plot_age_series(
col = col,
y_lab = y_lab,
line_width = 2,
boundary = 0.5,
point_size = 1,
error_bar_width = 0.5,
error_bar_linewidth = 0.5,
error_bar = TRUE,
text_size = env_vars$base_size
)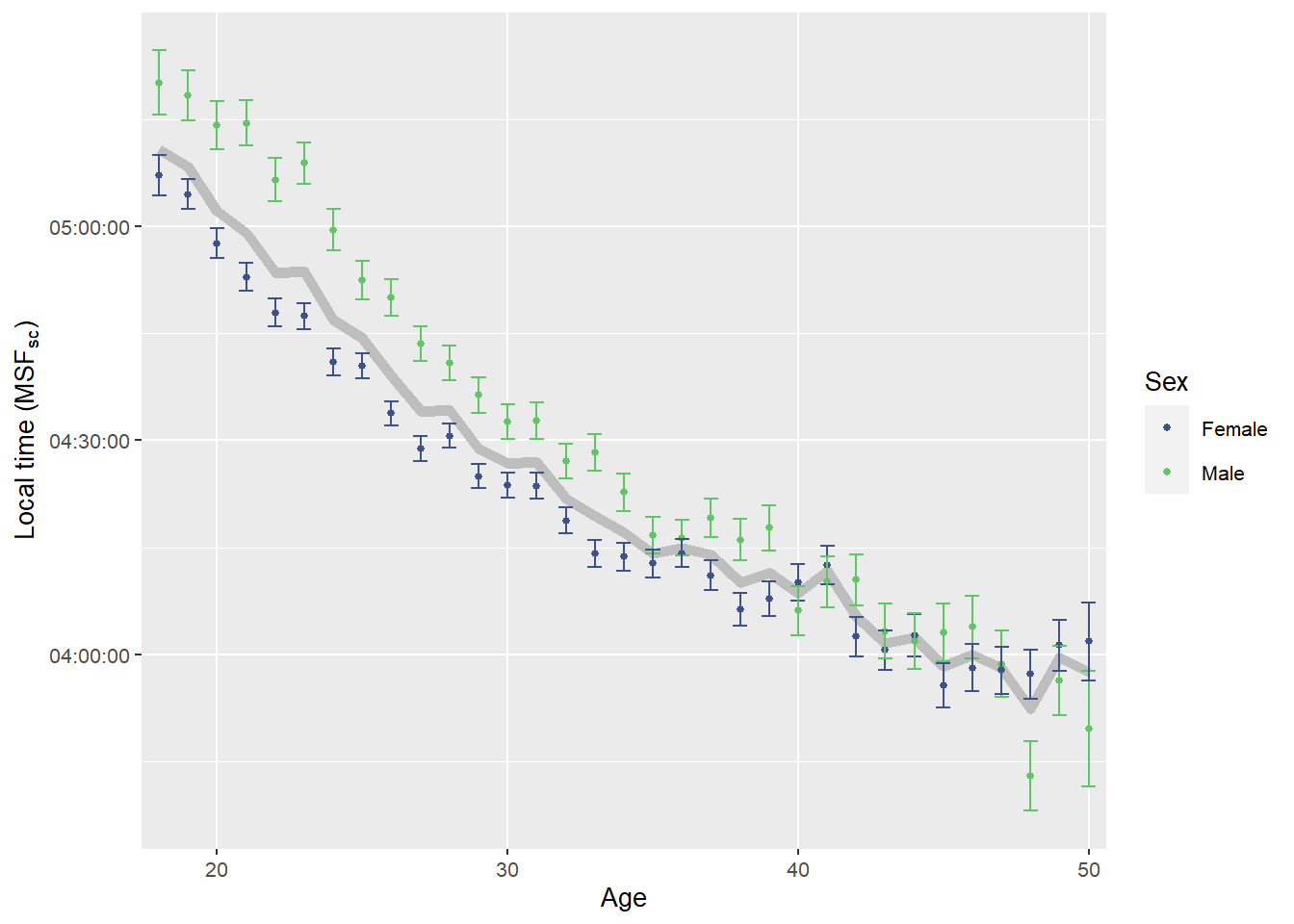
Source: Created by the author. Based on data visualization found in Roenneberg et al. (2007).
C.6.2 Age/sex versus weight
source(here::here("R/plot_age_series.R"))
col <- "weight"
y_lab <- "Weight (kg)"
data |>
dplyr::filter(age <= 50) |>
plot_age_series(
col = col,
y_lab = y_lab,
line_width = 2,
boundary = 0.5,
point_size = 1,
error_bar_width = 0.5,
error_bar_linewidth = 0.5,
error_bar = TRUE,
text_size = env_vars$base_size
)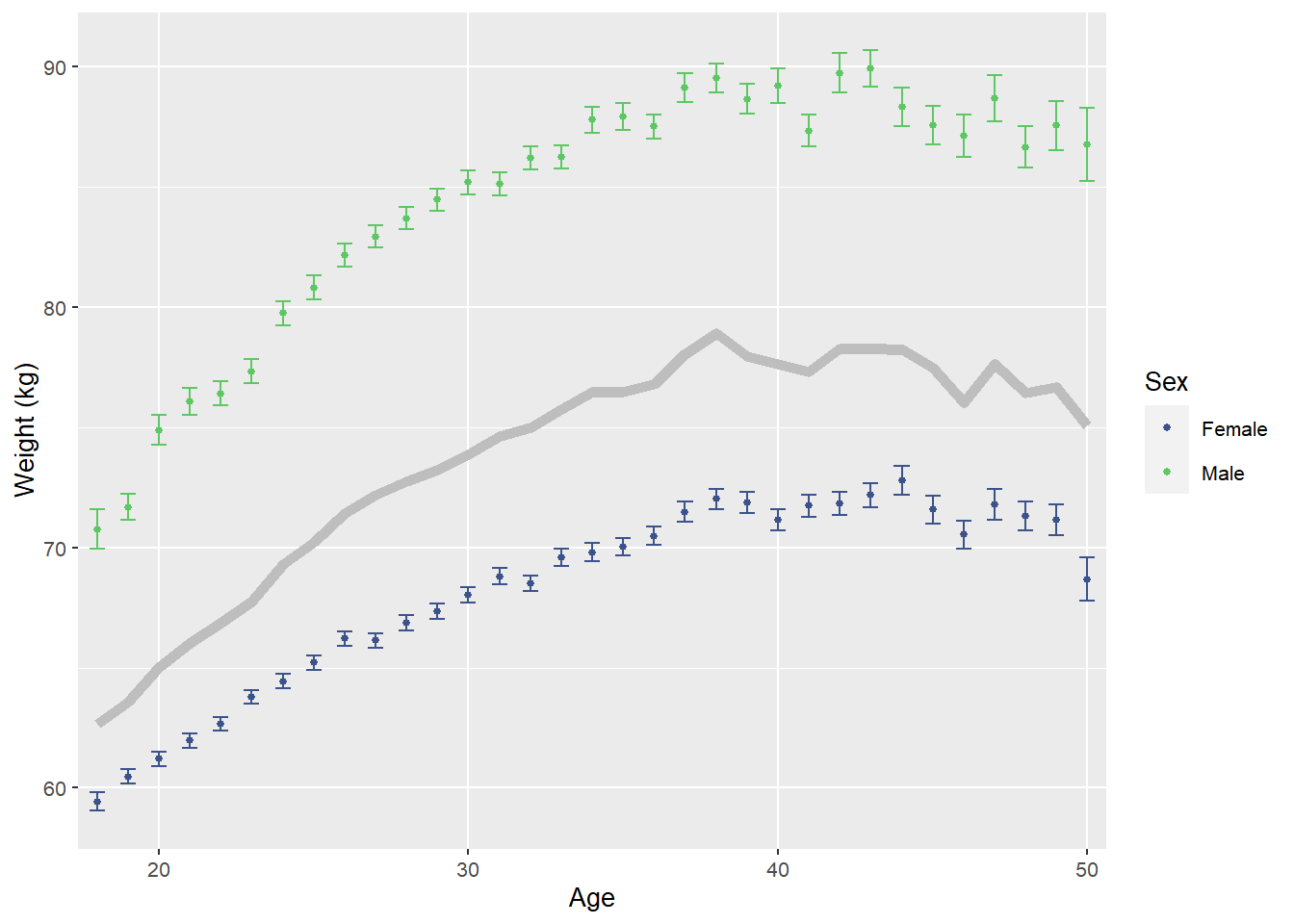
Source: Created by the author. Based on data visualization found in Roenneberg et al. (2007).
C.7 Chronotype distribution
source(here::here("R/plot_chronotype.R"))
col <- "msf_sc"
y_lab <- latex2exp::TeX("Local time ($MSF_{sc}$)")
data |>
plot_chronotype(
col = col,
x_lab = "Frequency (%)",
y_lab = y_lab,
col_width = 0.8,
col_border = 0.6,
text_size = env_vars$base_size,
legend_position = "right",
chronotype_cuts = FALSE
)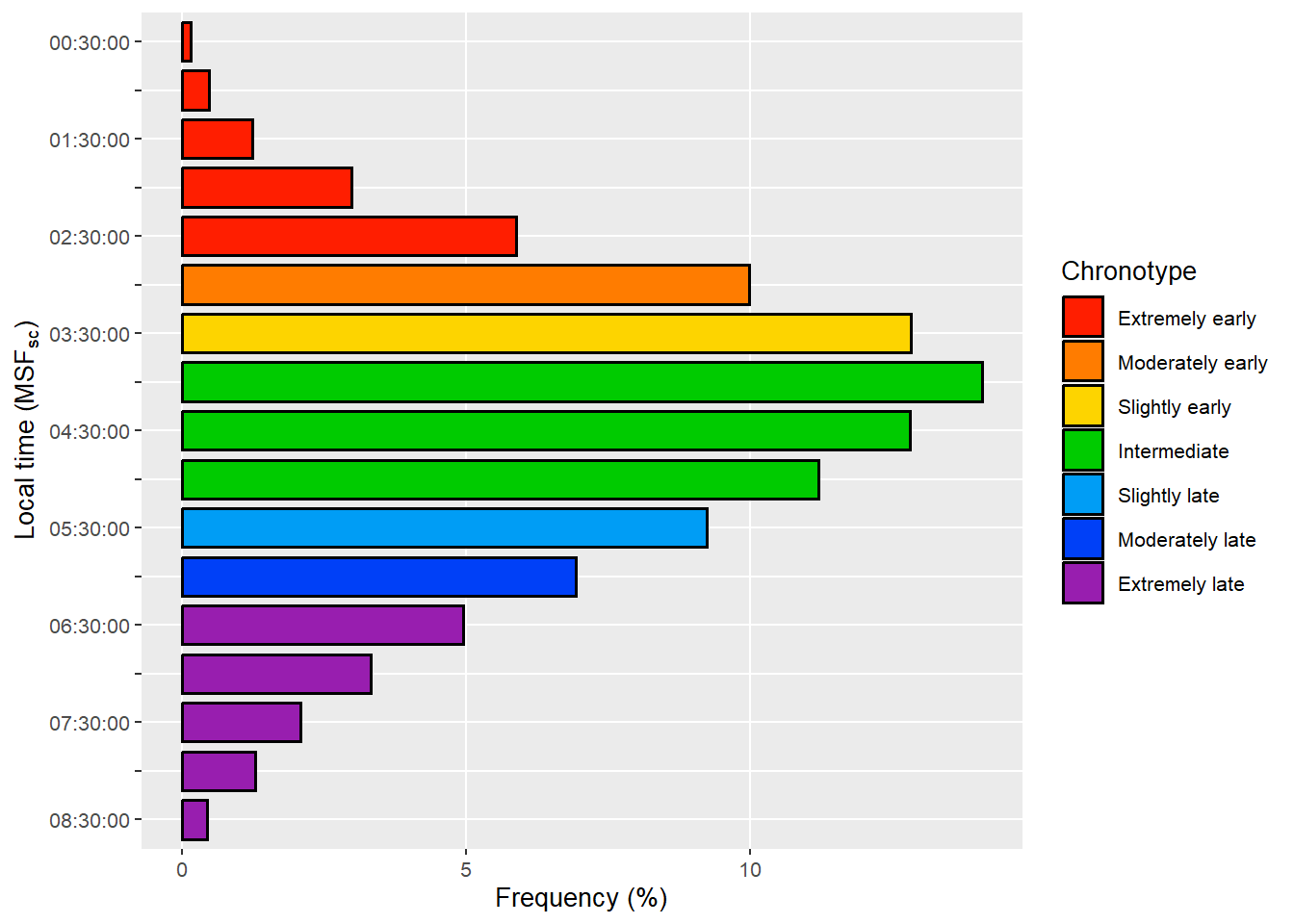
Source: Created by the author. Based on data visualization found in Roenneberg et al. (2019).
C.8 Latitude series
source(here::here("R/plot_latitude_series.R"))
col <- "msf_sc"
y_lab <- latex2exp::TeX("$MSF_{sc} \\pm SEM$")
data |>
dplyr::filter(age <= 50) |>
plot_latitude_series(
col = col,
y_lab = y_lab,
line_width = 2,
point_size = 3,
error_bar_width = 0.5,
error_bar_linewidth = 1,
error_bar = TRUE,
text_size = env_vars$base_size
)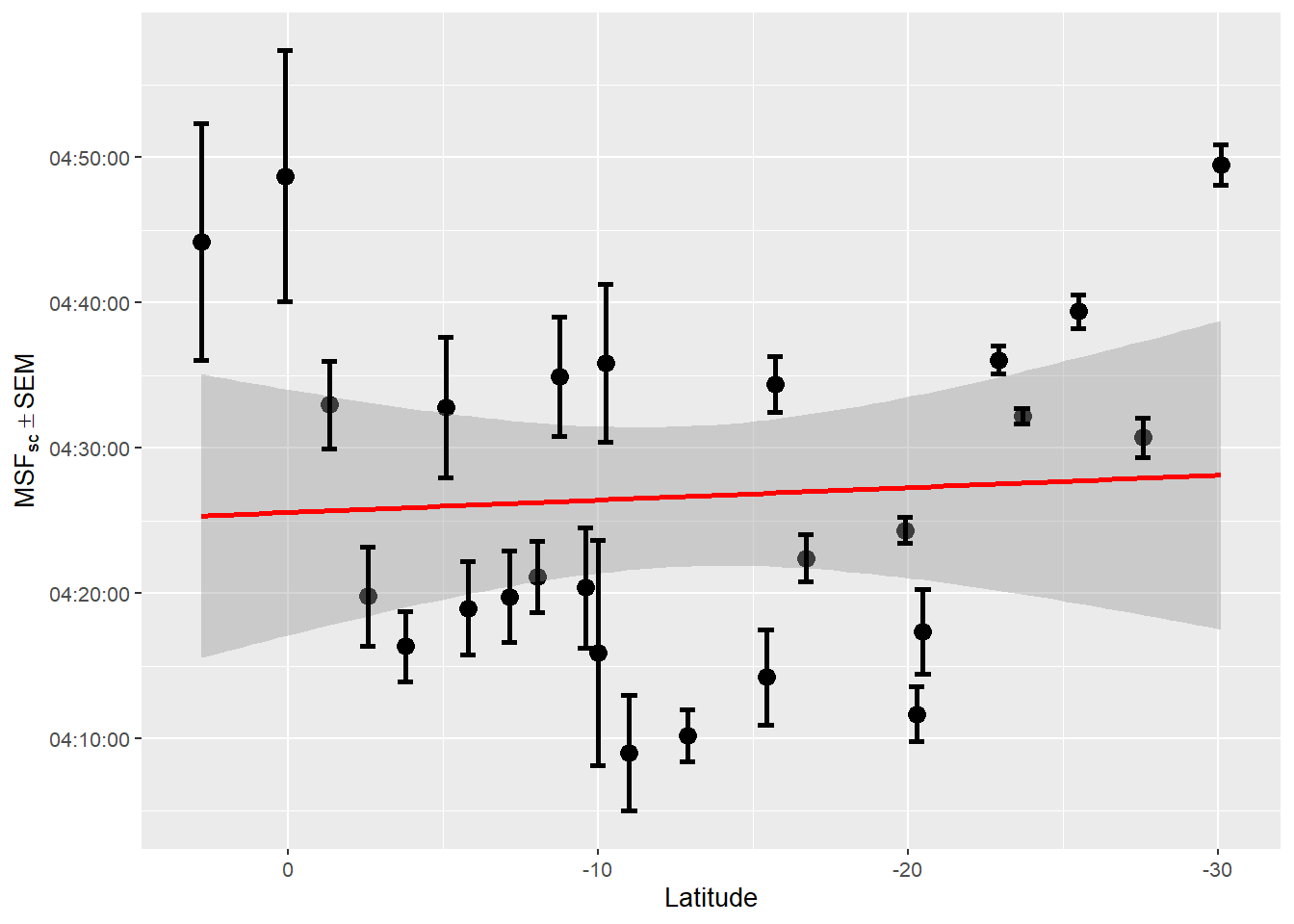
Source: Created by the author. Based on data visualization found in Leocadio-Miguel et al. (2017).
C.9 Statistics
C.9.1 Numerical variables
source(here::here("R/stats_sum.R"))
source(here::here("R/utils.R"))
col <- "msf_sc"
data |>
magrittr::extract2(col) |>
stats_sum(print = FALSE) |>
list_as_tibble()| name | value |
|---|---|
| n | 79198 |
| n_rm_na | 79198 |
| n_na | 0 |
| mean | 04:28:17.770957 |
| var | 08:05:53.49992 |
| sd | 01:26:51.406096 |
| min | 00:22:30 |
| q_1 | 03:26:25.714286 |
| median | 04:20:42.857143 |
| q_3 | 05:25:42.857143 |
| max | 08:31:04.285714 |
| iqr | 01:59:17.142857 |
| skewness | 0.284586184927996 |
| kurtosis | 2.77321491178072 |
Source: Created by the author.
C.9.2 Sex
# See <https://sidra.ibge.gov.br> to learn more.
library(magrittr)
rutils:::assert_internet()
# Brazil's 2022 census data
census_data <-
sidrar::get_sidra(x = 9514) %>% # Don't change the pipe
dplyr::filter(
Sexo %in% c("Homens", "Mulheres", "Total"),
stringr::str_detect(
Idade,
"^(1[8-9]|[2-9][0-9]) (ano|anos)$|^100 anos ou mais$"
),
.[[17]] == "Total"
) |>
dplyr::transmute(
sex = dplyr::case_when(
Sexo == "Homens" ~ "Male",
Sexo == "Mulheres" ~ "Female",
Sexo == "Total" ~ "Total"
),
value = Valor
) |>
dplyr::group_by(sex) |>
dplyr::summarise(n = sum(value)) |>
dplyr::ungroup()
census_data <-
dplyr::bind_rows(
census_data |>
dplyr::filter(sex != "Total") |>
dplyr::mutate(
n_rel = n / sum(n[sex != "Total"]),
n_per = round(n_rel * 100, 3)
),
census_data |>
dplyr::filter(sex == "Total") |>
dplyr::mutate(n_rel = 1, n_per = 100)
) |>
dplyr::as_tibble() |>
dplyr::arrange(sex)
count <- data |>
dplyr::select(sex) |>
dplyr::group_by(sex) |>
dplyr::summarise(n = dplyr::n()) |>
dplyr::ungroup() |>
dplyr::mutate(
n_rel = n / sum(n),
n_per = round(n_rel * 100, 3)
) |>
dplyr::arrange(dplyr::desc(n_rel)) |>
dplyr::bind_rows(
dplyr::tibble(
sex = "Total",
n = nrow(tidyr::drop_na(data, sex)),
n_rel = 1,
n_per = 100
)
)
count <-
dplyr::left_join(
count, census_data,
by = "sex",
suffix = c("_sample", "_census")
) |>
dplyr::mutate(
n_rel_diff = n_rel_sample - n_rel_census,
n_per_diff = n_per_sample - n_per_census
) |>
dplyr::relocate(
sex, n_sample, n_census, n_rel_sample, n_rel_census, n_rel_diff,
n_per_sample, n_per_census, n_per_diff
)
count |> dplyr::select(sex, n_per_sample, n_per_census, n_per_diff)| sex | n_per_sample | n_per_census | n_per_diff |
|---|---|---|---|
| Female | 66.243 | 52.263 | 13.98 |
| Male | 33.757 | 47.737 | -13.98 |
| Total | 100.000 | 100.000 | 0.00 |
Source: Created by the author. Based on data from Brazil’s 2022 census (Instituto Brasileiro de Geografia e Estatística (n.d.-b)).
sum(count$n_per_diff)
#> [1] -7.105427e-15C.9.3 Age and sex
source(here::here("R/stats_sum.R"))
source(here::here("R/utils.R"))
value <- "Male"
data |>
dplyr::filter(sex == value) |>
magrittr::extract2("age") |>
stats_sum(print = FALSE) |>
list_as_tibble()| name | value |
|---|---|
| n | 26735 |
| n_rm_na | 26735 |
| n_na | 0 |
| mean | 32.4343759740665 |
| var | 80.9906211885464 |
| sd | 8.99947893983571 |
| min | 18 |
| q_1 | 25.5388888888889 |
| median | 31.2583333333333 |
| q_3 | 37.9319444444444 |
| max | 58.7722222222222 |
| iqr | 12.3930555555556 |
| skewness | 0.617696405622681 |
| kurtosis | 2.84390555184727 |
Source: Created by the author.
# See <https://sidra.ibge.gov.br> to learn more.
library(magrittr)
rutils:::assert_internet()
# Brazil's 2022 census data
census_data <-
sidrar::get_sidra(x = 9514) %>% # Don't change the pipe
dplyr::filter(
Sexo %in% c("Homens", "Mulheres", "Total"),
stringr::str_detect(
Idade,
"^(1[8-9]|[2-9][0-9]) (ano|anos)$|^100 anos ou mais$"
),
.[[17]] == "Total"
) |>
dplyr::transmute(
sex = dplyr::case_when(
Sexo == "Homens" ~ "Male",
Sexo == "Mulheres" ~ "Female",
Sexo == "Total" ~ "Total"
),
age = as.numeric(stringr::str_extract(Idade, "\\d+")),
value = Valor
) |>
dplyr::group_by(sex) |>
dplyr::summarise(
mean = stats::weighted.mean(age, value),
sd = sqrt(Hmisc::wtd.var(age, value))
) |>
dplyr::ungroup() |>
dplyr::mutate(
min = c(18, 18, 18),
max = c(100, 100, 100)
) |>
dplyr::relocate(sex, mean, sd, min, max) |>
dplyr::as_tibble()
count <- data |>
dplyr::select(sex, age) |>
dplyr::group_by(sex) |>
dplyr::mutate(sex = as.character(sex)) |>
dplyr::summarise(
mean = mean(age, na.rm = TRUE),
sd = stats::sd(age, na.rm = TRUE),
min = min(age, na.rm = TRUE),
max = max(age, na.rm = TRUE)
) |>
dplyr::ungroup() |>
dplyr::bind_rows(
dplyr::tibble(
sex = "Total",
mean = mean(data$age, na.rm = TRUE),
sd = stats::sd(data$age, na.rm = TRUE),
min = min(data$age, na.rm = TRUE),
max = max(data$age, na.rm = TRUE)
)
)
count <-
dplyr::left_join(
count,
census_data,
by = "sex",
suffix = c("_sample", "_census")
) |>
dplyr::mutate(mean_diff = mean_sample - mean_census) |>
dplyr::relocate(
sex, mean_sample, mean_census, mean_diff, sd_sample, sd_census,
min_sample, min_census, max_sample, max_census
)
count |>
dplyr::select(
sex, mean_sample, mean_census, mean_diff, sd_sample, sd_census
)| sex | mean_sample | mean_census | mean_diff | sd_sample | sd_census |
|---|---|---|---|---|---|
| Female | 31.75420 | 44.98722 | -13.23302 | 9.340939 | 17.51132 |
| Male | 32.43438 | 43.49903 | -11.06465 | 8.999479 | 16.86385 |
| Total | 31.98381 | 44.27680 | -12.29299 | 9.232632 | 17.22133 |
Source: Created by the author. Based on data from Brazil’s 2022 census (Instituto Brasileiro de Geografia e Estatística (n.d.-b)).
sum(count$mean_diff)
#> [1] -36.59066C.9.4 Longitudinal range
C.9.4.1 Sample
source(here::here("R/stats_sum.R"))
source(here::here("R/utils.R"))
stats <-
data |>
magrittr::extract2("longitude") |>
stats_sum(print = FALSE)
abs(stats$max - stats$min)
#> [1] 33.023
stats |> list_as_tibble()| name | value |
|---|---|
| n | 79198 |
| n_rm_na | 79198 |
| n_na | 0 |
| mean | -45.9455401815147 |
| var | 18.9406905927715 |
| sd | 4.35209037047388 |
| min | -67.9869962 |
| q_1 | -48.4296364 |
| median | -46.9249578 |
| q_3 | -43.7756411 |
| max | -34.9639996 |
| iqr | 4.6539953 |
| skewness | 0.0156480710174436 |
| kurtosis | 5.78918700160139 |
Source: Created by the author.
C.9.4.2 Brazil
change_sign <- function(x) x * (-1)
## Ponta do Seixas, PB (7° 09′ 18″ S, 34° 47′ 34″ O)
min <-
measurements::conv_unit("34 47 34", from = "deg_min_sec", to = "dec_deg") |>
as.numeric() |>
change_sign()
## Nascente do rio Moa, AC (7° 32′ 09″ S, 73° 59′ 26″ O)
max <-
measurements::conv_unit("73 59 26", from = "deg_min_sec", to = "dec_deg") |>
as.numeric() |>
change_sign()
min
#> [1] -34.79278
max
#> [1] -73.99056
abs(max - min)
#> [1] 39.19778C.9.5 Latitudinal range
C.9.5.1 Sample
source(here::here("R/stats_sum.R"))
source(here::here("R/utils.R"))
stats <-
data |>
magrittr::extract2("latitude") |>
stats_sum(print = FALSE)
abs(stats$max - stats$min)
#> [1] 32.91596
stats |> list_as_tibble()| name | value |
|---|---|
| n | 79198 |
| n_rm_na | 79198 |
| n_na | 0 |
| mean | -20.8338507528991 |
| var | 40.2956396934244 |
| sd | 6.34788466289554 |
| min | -30.1087672 |
| q_1 | -23.6820636 |
| median | -23.6820636 |
| q_3 | -19.9026404 |
| max | 2.8071961 |
| iqr | 3.7794232 |
| skewness | 1.40629570823769 |
| kurtosis | 4.67433697579443 |
Source: Created by the author.
C.9.5.2 Brazil
change_sign <- function(x) x * (-1)
## Arroio Chuí, RS (33° 45′ 07″ S, 53° 23′ 50″ O)
min <-
measurements::conv_unit("33 45 07", from = "deg_min_sec", to = "dec_deg") |>
as.numeric() |>
change_sign()
## Nascente do rio Ailã, RR (5° 16′ 19″ N, 60° 12′ 45″ O)
max <-
measurements::conv_unit("5 16 19", from = "deg_min_sec", to = "dec_deg") |>
as.numeric()
min
#> [1] -33.75194
max
#> [1] 5.271944
abs(max - min)
#> [1] 39.02389C.9.6 Region
# See <https://sidra.ibge.gov.br> to learn more.
rutils:::assert_internet()
# Brazil's 2022 census data
census_data <-
sidrar::get_sidra(x = 4714, variable = 93, geo = "Region") |>
dplyr::select(dplyr::all_of(c("Valor", "Grande Região"))) |>
dplyr::transmute(
col = `Grande Região`,
n = Valor,
n_rel = n / sum(n),
n_per = round(n_rel * 100, 3)
) |>
dplyr::mutate(
col = dplyr::case_when(
col == "Norte" ~ "North",
col == "Nordeste" ~ "Northeast",
col == "Centro-Oeste" ~ "Midwest",
col == "Sudeste" ~ "Southeast",
col == "Sul" ~ "South"
)
) |>
dplyr::as_tibble() |>
dplyr::arrange(dplyr::desc(n_rel))
count <- data |>
magrittr::extract2("region") |>
stats_sum(print = FALSE) |>
magrittr::extract2("count") |>
dplyr::mutate(
n_rel = n / sum(n),
n_per = round(n_rel * 100, 3)
) |>
dplyr::arrange(dplyr::desc(n_rel))
count <-
dplyr::left_join(
count, census_data, by = "col", suffix = c("_sample", "_census")
) |>
dplyr::mutate(
n_rel_diff = n_rel_sample - n_rel_census,
n_per_diff = n_per_sample - n_per_census
) |>
dplyr::relocate(
col, n_sample, n_census, n_rel_sample, n_rel_census, n_rel_diff,
n_per_sample, n_per_census, n_per_diff
)
count |> dplyr::select(col, n_per_sample, n_per_census, n_per_diff)| col | n_per_sample | n_per_census | n_per_diff |
|---|---|---|---|
| Southeast | 60.565 | 41.777 | 18.788 |
| South | 17.122 | 14.742 | 2.380 |
| Northeast | 11.538 | 26.914 | -15.376 |
| Midwest | 8.287 | 8.021 | 0.266 |
| North | 2.489 | 8.546 | -6.057 |
Source: Created by the author. Based on data from Brazil’s 2022 census (Instituto Brasileiro de Geografia e Estatística (n.d.-a)).
sum(count$n_per_diff)
#> [1] 0.001C.9.7 State
source(here::here("R/stats_sum.R"))
data |>
magrittr::extract2("state") |>
stats_sum(print = FALSE) |>
magrittr::extract2("count") |>
dplyr::mutate(
n_rel = n / sum(n),
n_per = round(n_rel * 100, 3)
) |>
dplyr::arrange(dplyr::desc(n_rel))| col | n | n_rel | n_per |
|---|---|---|---|
| São Paulo | 26379 | 0.3330766 | 33.308 |
| Minas Gerais | 10115 | 0.1277179 | 12.772 |
| Rio de Janeiro | 9381 | 0.1184500 | 11.845 |
| Paraná | 5517 | 0.0696609 | 6.966 |
| Rio Grande do Sul | 4097 | 0.0517311 | 5.173 |
| Santa Catarina | 3946 | 0.0498245 | 4.982 |
| Goiás | 2674 | 0.0337635 | 3.376 |
| Bahia | 2522 | 0.0318442 | 3.184 |
| Espírito Santo | 2091 | 0.0264022 | 2.640 |
| Distrito Federal | 2087 | 0.0263517 | 2.635 |
| Pernambuco | 1550 | 0.0195712 | 1.957 |
| Ceará | 1398 | 0.0176520 | 1.765 |
| Mato Grosso do Sul | 1014 | 0.0128034 | 1.280 |
| Pará | 938 | 0.0118437 | 1.184 |
| Rio Grande do Norte | 789 | 0.0099624 | 0.996 |
| Mato Grosso | 788 | 0.0099497 | 0.995 |
| Paraíba | 773 | 0.0097603 | 0.976 |
| Maranhão | 652 | 0.0082325 | 0.823 |
| Sergipe | 533 | 0.0067300 | 0.673 |
| Alagoas | 526 | 0.0066416 | 0.664 |
| Rondônia | 401 | 0.0050633 | 0.506 |
| Piauí | 395 | 0.0049875 | 0.499 |
| Tocantins | 268 | 0.0033839 | 0.338 |
| Acre | 132 | 0.0016667 | 0.167 |
| Roraima | 119 | 0.0015026 | 0.150 |
| Amapá | 113 | 0.0014268 | 0.143 |
Source: Created by the author. Based on data from Brazil’s 2022 census (Instituto Brasileiro de Geografia e Estatística (n.d.-a)).